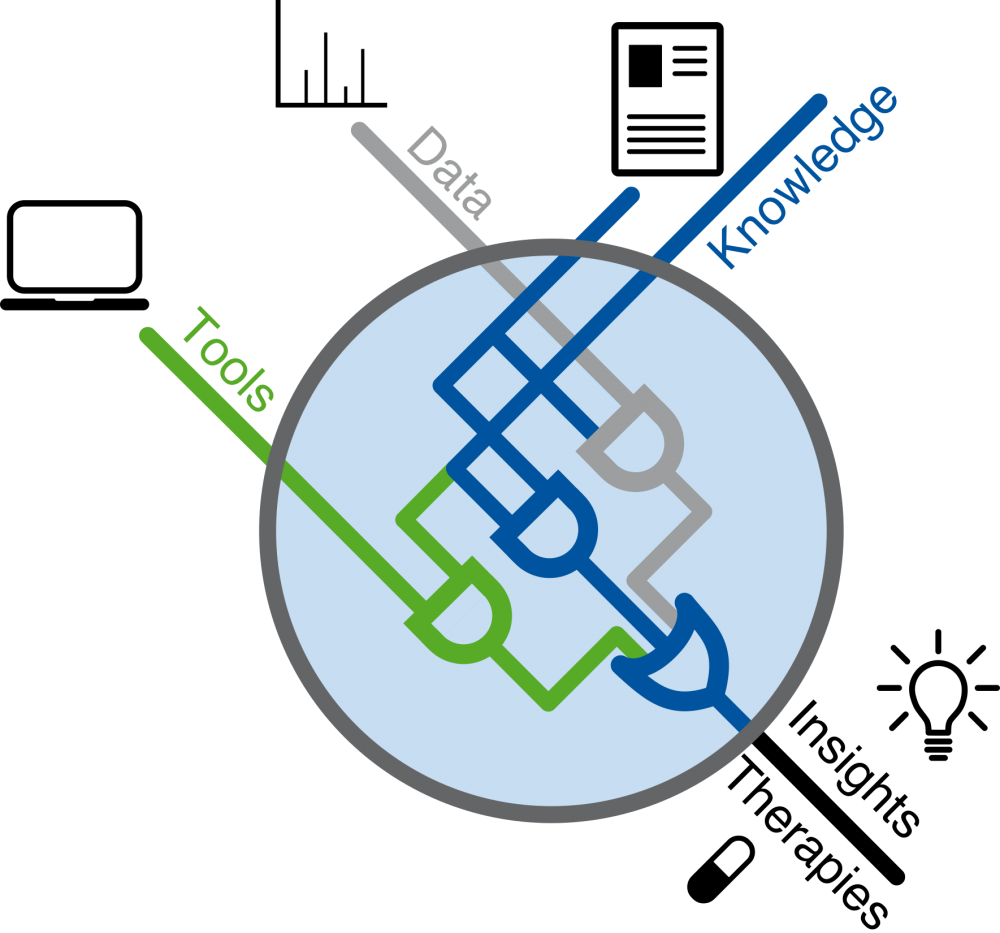
Our goal is to acquire a functional understanding of the deregulation of signalling networks in disease and to apply this knowledge to develop novel therapeutics. We focus on cancer, auto-immune and fibrotic disease. Towards this goal, we integrate big (‘Omics’) data with mechanistic molecular knowledge into statistical and machine learning methods, and we share our tools as free open-source packages.
Our research is application-driven and tailored towards producing computational models that integrate diverse data sources to better understand and treat diseases. Because of this, we collaborate closely with experimental groups. A major emphasis is to build context-specific models that are both mechanistic (to provide understanding) and predictive (to generate novel hypotheses). To build these models, we combine existing biochemical knowledge with different types of large scale data. We believe that this biological knowledge can be instrumental to move from pure correlation to causation in large data sets, and thereby identify the molecular processes that underlie specific phenomena.
We develop and apply methods to extract mechanistic features from diverse omics data, recently also for single-cell data. We then combine these multi-omics data sets into causal networks. Finally, we build dynamic models of specific subsystems using logic formalisms that we can analyze and simulate to predict the effect of new perturbations.
We apply these strategies in the context of many disease conditions. Particular areas of interest for us are:
- Cancer
- Auto-immune diseases
- Fibrotic diseases
While our research is driven by applications, we develop open-source computational tools that share freely with the scientific community.
Finally, we support scientific crowdsourcing, specifically collaborative competitions, through the DREAM challenges.
We are at the European Bioinformatics Institute (EMBL-EBI) and at the Institute for Computational Biomedicine at the Medical Faculty of Heidelberg University and Heidelberg University Hospital. We are also part of ELLIS Heidelberg.